"""Implementation of evolution strategy for gray box coefficient estimation."""
import numpy as np
import numpy.random as rnd
import matplotlib.pyplot as plt
def f(a, b, c, x):
"""Return the value of the function to be estimated (the gray box)."""
return a*x**2 + b*np.cos(c*x*np.pi)
def initial_x():
"""Return the vector of initial estimated coefficient."""
a = min(max(rnd.randn()*5, -10), 10)
b = min(max(rnd.randn()*5, -10), 10)
c = min(max(rnd.randn()*5, -10), 10)
return [a, b, c]
def initial_sigma():
"""Return the vector of initial mutation factors."""
s_a = rnd.randn()
s_b = rnd.randn()
s_c = rnd.randn()
return [s_a, s_b, s_c]
class Population:
"""Class for managing the evolving population."""
def __init__(self, pop_size=100, mutations_count=5):
"""Initialize a population and neccessary inner values."""
# The population holding vectors of solutions and their mutations
self.population = [initial_x() + initial_sigma()
for i in range(pop_size)]
# The maximal size of the population
self.pop_size = pop_size
# The amount of offspring each parent has
self.mutations_count = mutations_count
# Mutations coefficients
self.tau_1 = 1/np.sqrt(2*pop_size)
self.tau_2 = 1/np.sqrt(2*np.sqrt(pop_size))
def load_data(self, dataset_num):
"""Load x and y data from the sspecified dataset."""
data = np.loadtxt('./data/ES_data_' + str(dataset_num) + '.dat')
self.data_x = [datapoint[0] for datapoint in data]
self.data_y = [datapoint[1] for datapoint in data]
def mutate(self):
"""Produce offspring for every vector in population."""
offspring = []
for mutation in range(self.mutations_count):
mutated = []
for i in range(self.pop_size):
# Calculate first mutation-changing coefficient
rnd_tau = rnd.randn() * self.tau_1
# Get a vector of target values and their mutations
v = np.array(self.population[i])
# Update target values using their mutations
a = v[0] + rnd.randn() * v[3]
b = v[1] + rnd.randn() * v[4]
c = v[2] + rnd.randn() * v[5]
# Update mutations using mutation coefficients
s_a = v[3] * np.exp(rnd_tau) * np.exp(rnd.randn()*self.tau_2)
s_b = v[4] * np.exp(rnd_tau) * np.exp(rnd.randn()*self.tau_2)
s_c = v[5] * np.exp(rnd_tau) * np.exp(rnd.randn()*self.tau_2)
# Add each newly created vector to the mutated population
mutated.append([a, b, c, s_a, s_b, s_c])
# Combine the mutated populations and return them as the offspring
offspring += mutated
return offspring
def evaluate(self, group, best=False, indices=False):
"""Return a dictionary of vector indices and theirs errors in order."""
# Create and populate the dictionary
error = {}
for i in range(len(group)):
v = np.array(group[i])
estimated_y = [f(v[0], v[1], v[2], x) for x in self.data_x]
error[i] = np.sum([(estimated_y[i] - self.data_y[i])**2
for i in range(len(self.data_y))])
# Sort the dictionary by values
error = {index: error for (index, error) in sorted(error.items(),
key=lambda pair: pair[1])}
if best:
return list(error.values())[0]
if indices:
return list(error.keys())
return error
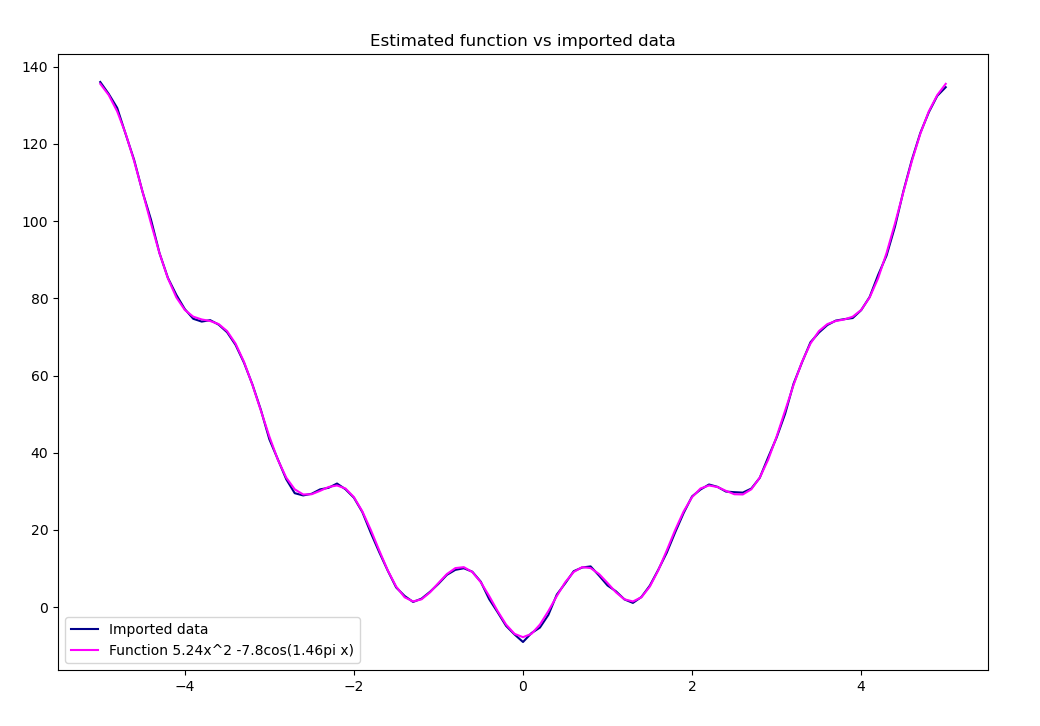
def plot_results(self, a, b, c):
"""Plot estimated and measured values."""
estimated_y = [f(a, b, c, x) for x in self.data_x]
errors = [estimated_y[i] - self.data_y[i]
for i in range(len(self.data_y))]
# Plot the data and estimated function on one plot
plt.figure(figsize=(12, 8))
plt.plot(self.data_x, self.data_y, color='darkblue')
plt.plot(self.data_x, estimated_y, color='magenta')
if b < 0:
plt.legend(['Imported data', f'Function {round(a, 2)}x^2 '
+ f'{round(b, 2)}cos({round(c, 2)}pi x)'])
else:
plt.legend(['Imported data', f'Function {round(a, 2)}x^2 + '
+ f'{round(b, 2)}cos({round(c, 2)}pi x)'])
plt.title('Estimated function vs imported data')
plt.show()
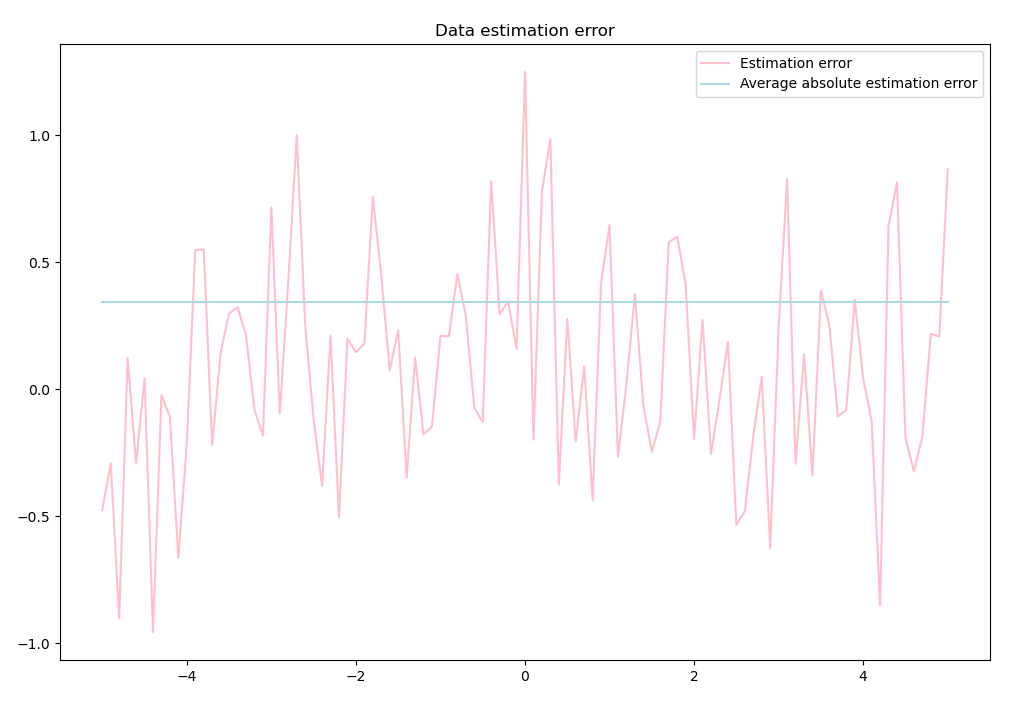
# Plot estimation errors
plt.figure(figsize=(12, 8))
plt.plot(self.data_x, errors, color='pink')
plt.plot(self.data_x, [np.mean(list(map(abs, errors)))
for _ in self.data_x], color='lightblue')
plt.legend(['Estimation error', 'Average absolute estimation error'])
plt.title('Data estimation error')
plt.show()
if __name__ == '__main__':
# Initialize the population and load a specific dataset to approximate
p = Population()
p.load_data(19)
iterations = 0
# State wanted accuracy (difference between best parent and best offspring)
eps = 1e-3
while(True):
# Perform mutation, combination and reduction to the original size
offspring = p.mutate()
best_parent = p.evaluate(p.population, best=True)
best_offspring = p.evaluate(offspring, best=True)
p.population = p.population + offspring
p.population = [p.population[i] for i in p.evaluate(
p.population, indices=True)[:p.pop_size]]
iterations += 1
# This difference of best individuals marks our stop condition
if abs(best_parent - best_offspring) < eps:
break
print(f'Condition met in {iterations} iterations')
# The best estimated coefficients
a, b, c = p.population[0][:3]
print(f'Estimated values:\na = {a}\nb = {b}\nc = {c}')
# Present the results with a graph
p.plot_results(a, b, c)